General Questions & Answers
We used GPT to evaluate research abstracts and identify pairs of entities (e.g., genes, proteins, metabolites)
in addition to their unique interaction type (e.g., enhances, binds, affects, represses).
For instance, after examining the abstract
"Protein-protein interactions of proteins from the ESAT-6 family of Mycobacterium tuberculosis. In the present study, we demonstrate that, in analogy with the genes encoding ESAT-6 and CFP-10, the genes rv0287 and rv0288 from the ESAT-6 gene family are cotranscribed. Using Western-Western blotting and protein-print overlay methodologies, we demonstrate that ESAT-6 and CFP-10, as well as the protein pair Rv0288/Rv0287, interact pairwise in a highly specific way. Most notably, the ESAT-6 proteins interact directly with Rv3873, a possible cell envelope component of the ESAT-6 secretion pathway."GPT returned the following statements:
Because the KnowledgeGraph database is built from processed abstracts and full-length texts, only standardized gene symbols that are used in literature can be used for searching.
You can also choose to search for terms mentioned in the abstracts (for instance, "cell membrane", "enzyme", "infection", etc.), author names, or PubMed IDs.
To ensure the KnowledgeGraph reflects state-of-the-art knowledge, all research papers were derived from PubMed and Elsevier. All articles related to the species gene information from various journals were extracted.
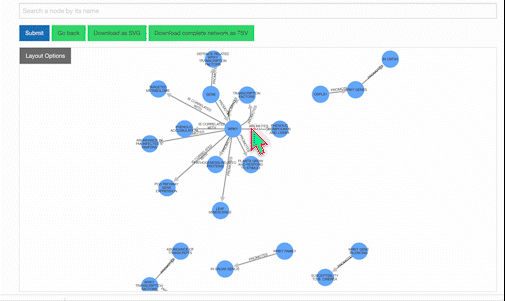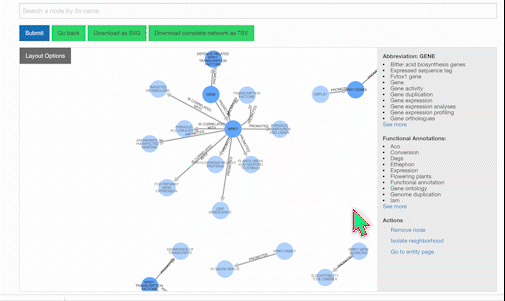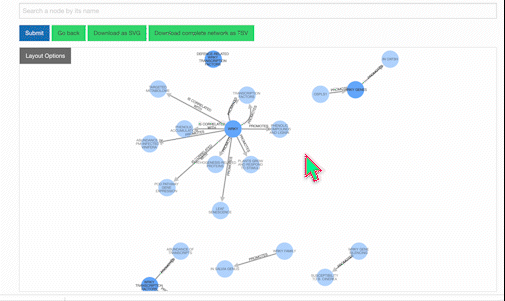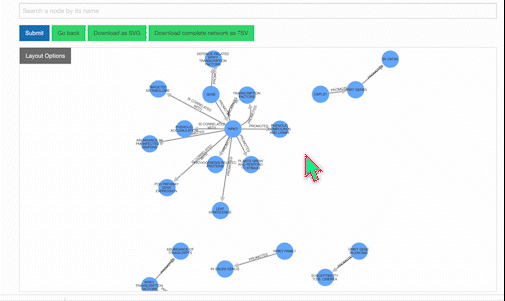
Our KnowledgeNetwork viewer displays a plethora of relationships found between your search query and GPT-detected entities. Should you want to narrow your search options, click on the "Layout Options" button:
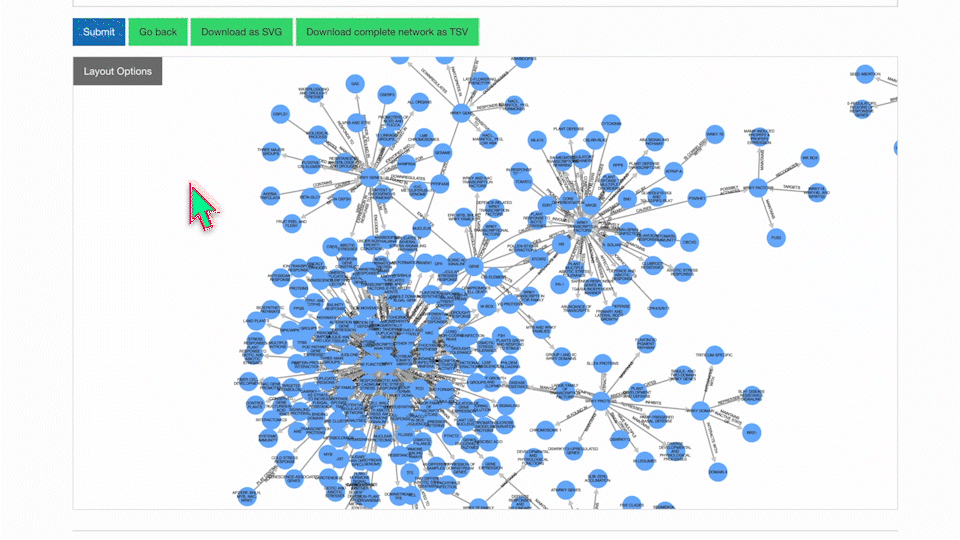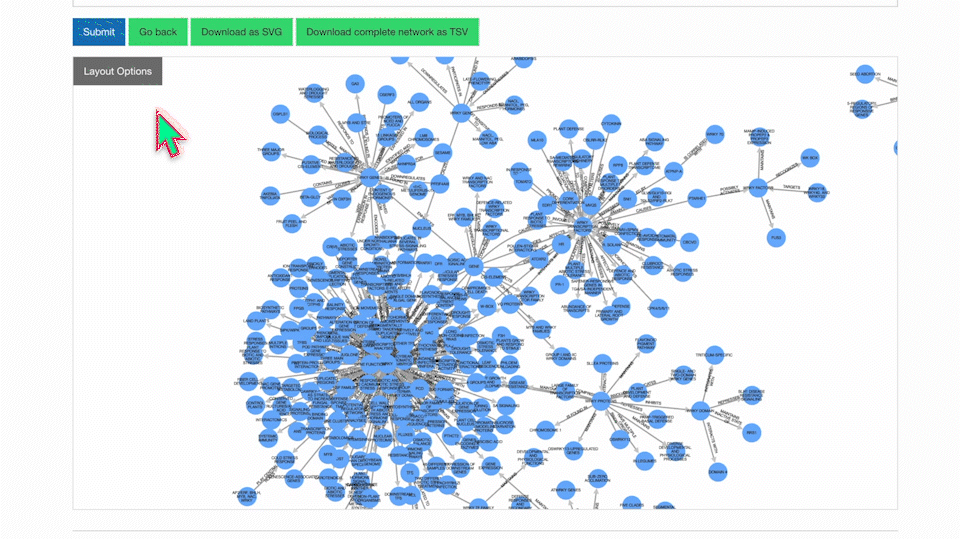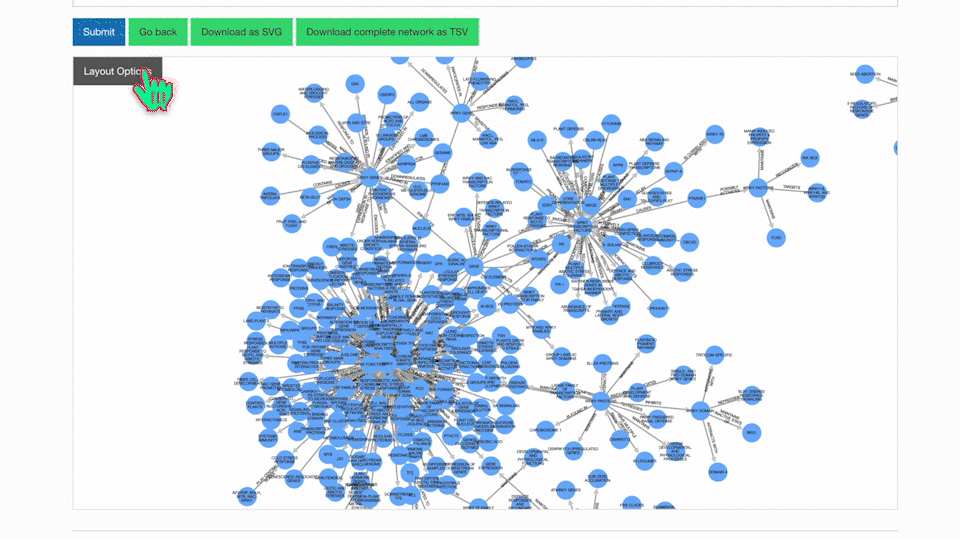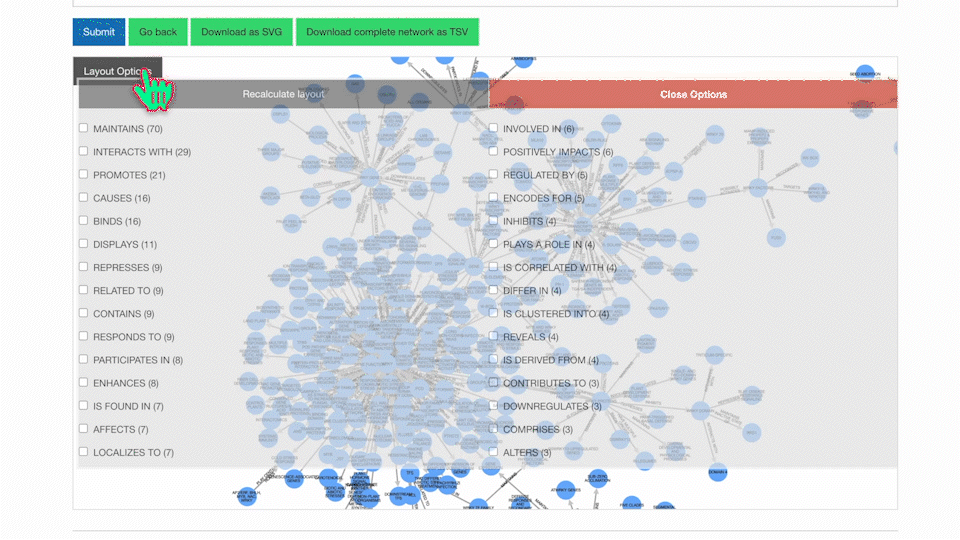
Then, select your edges of interest and click "Recalculate Layout":
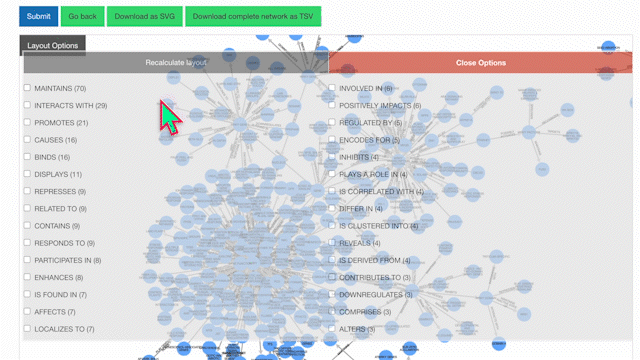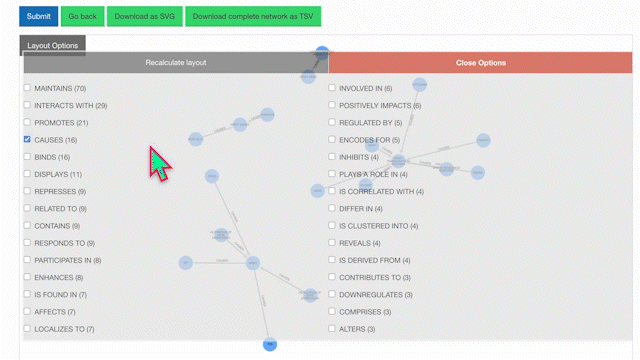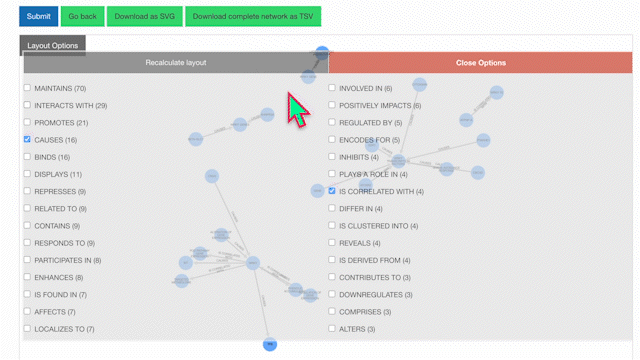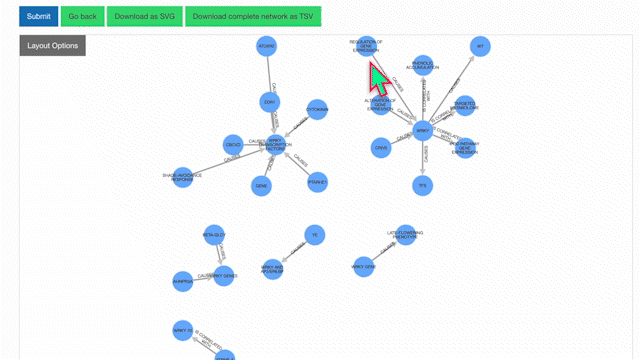
To learn more about a specific node, click on it:
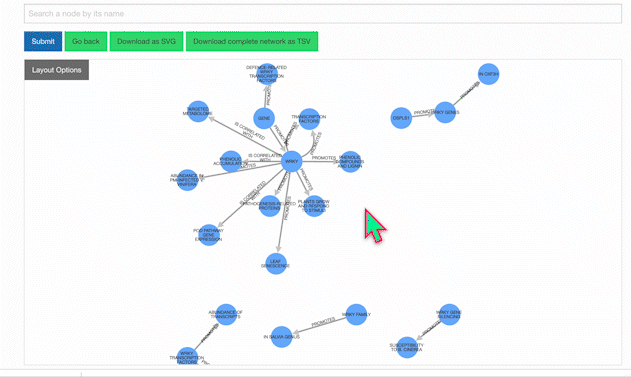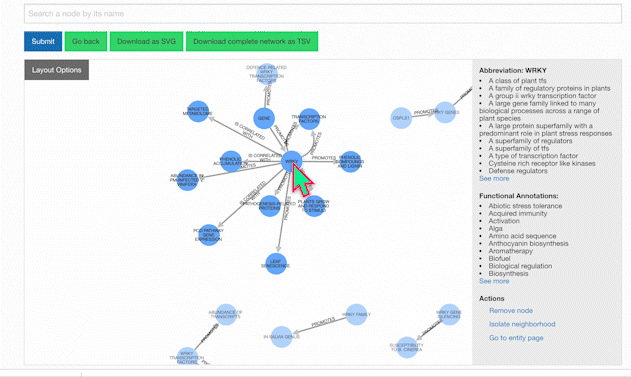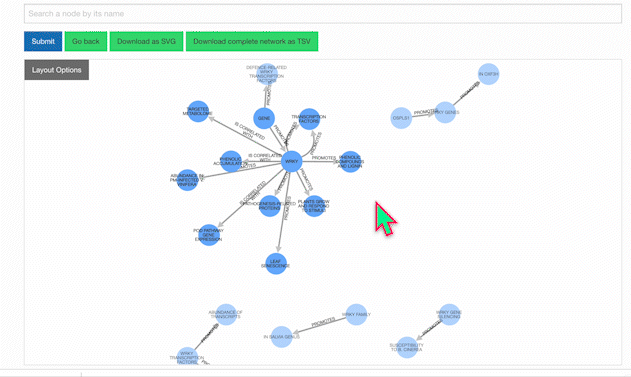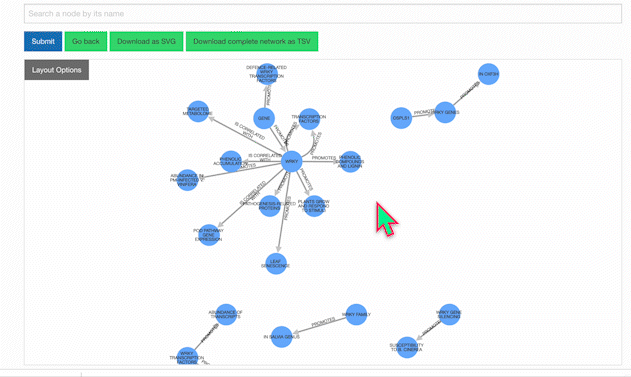
Users also have the option of personally modifying networks. For example, among the list of possible actions when clicking a node is removing it. Clicking once on the background will restore node opacity, while clicking twice will reincorporate the removed node.
You can use the TSVGraph script, which generates an interactive HTML file viewable in your web browser. To use it:
python cytoscape.py --file path/to/tsv-file --out path/to/output.html.path/to/tsv-file with the path to your TSV file and path/to/output.html with the desired output HTML file path.Below the KnowledgeNetwork viewer is a table showcasing the network's nodes and edges. Clicking on the Pubmed ID will display both the research abstract and entities identified from it:
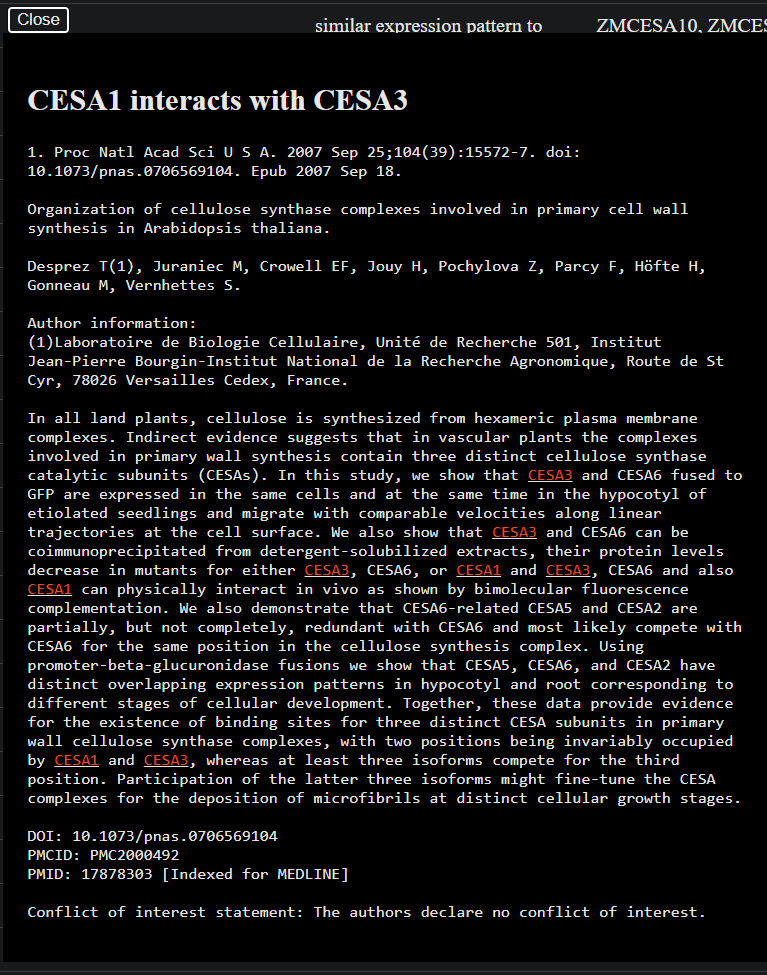
As with any AI model, GPT is not 100% accurate, generating results that may be erroneous or incomplete. Accordingly, relationships of interest warrant confirmation with the appropriate abstract. To estimate the accuracy rate, we sampled and manually inspected 50 articles, the results of which are summarized by the following 3 charts:
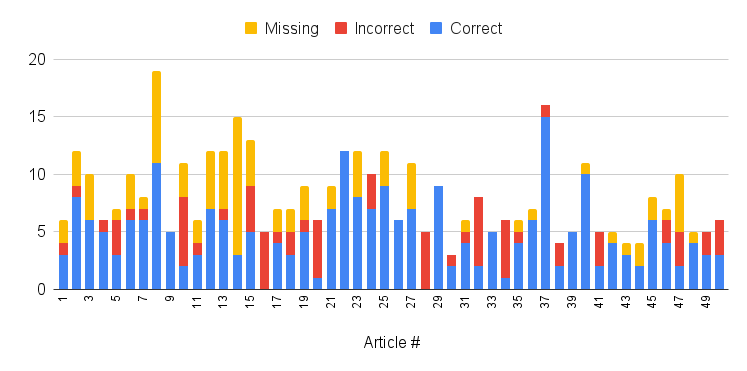
Distribution of correct, incorrect, and missing statements detected by GPT.
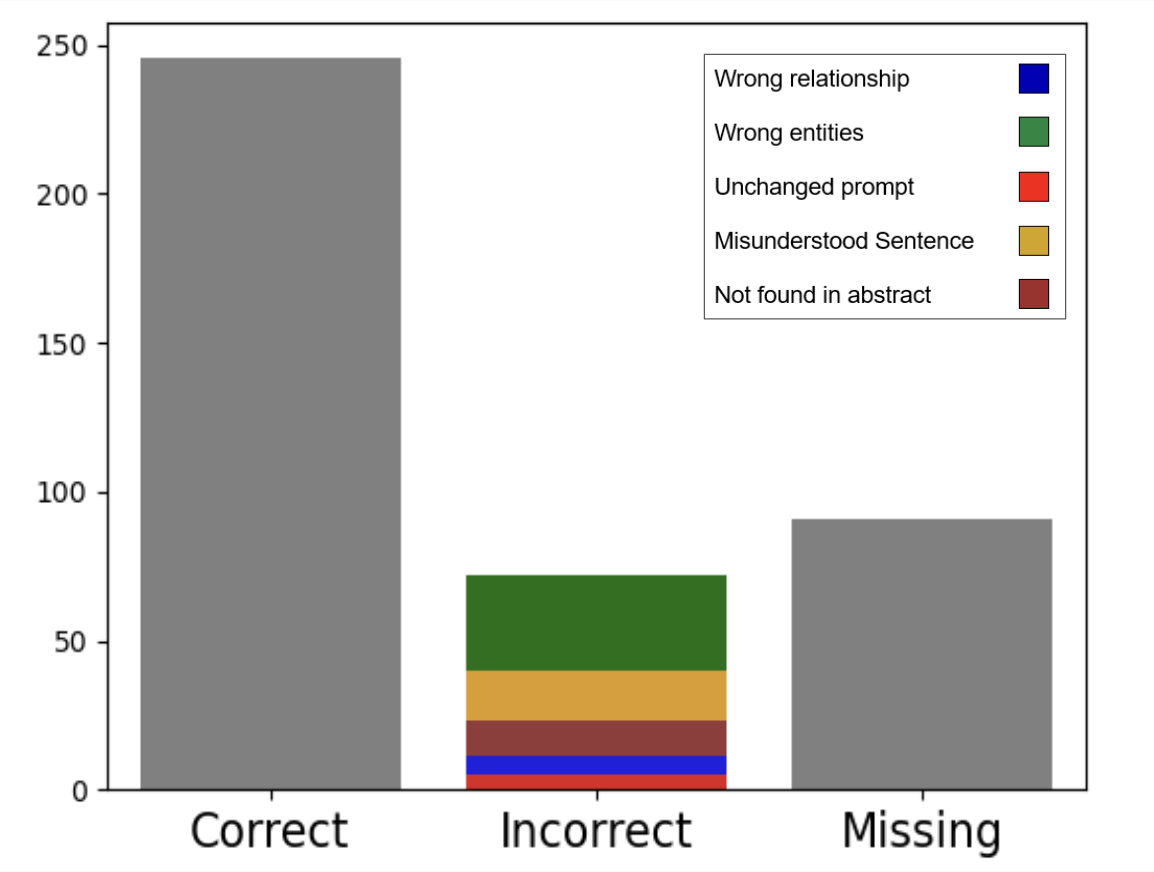
Total number of correct, incorrect, and missing statements extracted from the 50 articles.